Clinical Data Harmonization
Clinical and phenotypic data from each research study contains demographic and condition-specific information that ranges from common data types that allow for cross-cohort comparison, as well as study-specific terms that allow for deeper cohort exploration. Phenotypic information is harmonized using community- based ontologies and standards such as the Human Phenotype Ontology (HPO) and/or the NCI Thesaurus (NCIt). The goal of this harmonization is to allow for easier analysis across different datasets and disease types, both within the Kids First Data Resource Center as well as across other genomic datasets. The harmonization process is iterative with the goal of addressing scientific use cases. To highlight the scope of this endeavor, learn more: About the Research.
Genomic Harmonization
The goal of genomic harmonization is to provide an “analysis-ready” dataset that is “functionally equivalent” both across the Kids First datasets and other large-scale genomic data initiatives. As such, the initial pipelines are based on community best practices for joint genotyping of genetic variation. For pediatric cancer samples, we are currently investigating a number of somatic callers in and anticipate providing multiple pipelines for somatic calls. Structural variant, as well as RNA-seq based quantification, are on the near-term roadmap.
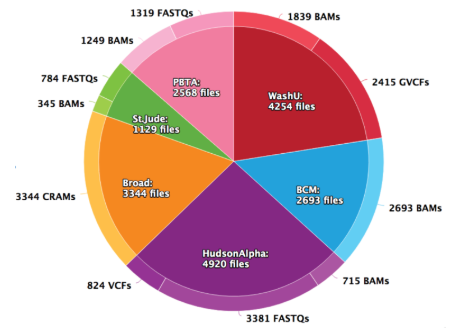
Data received by the DRC, spanning several years of data generation. Most of the data is whole genome sequencing of germline trio data, with RNA-seq and whole exome sequencing for tumor samples.
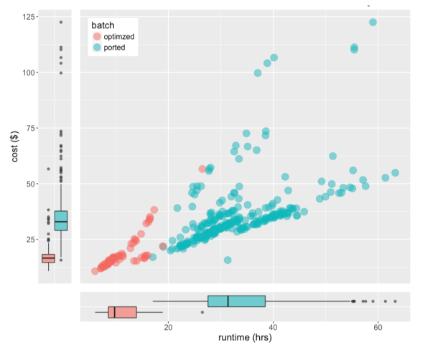
Cost and runtime analysis of whole genome alignment and gVCF generation running Cavatica using spot instances on AWS. The “ported” workflow was a standard port of BWA-mem and GATK workflows into CWL. The “optimized” workflow leveraged knowledge about the resources.